Biomanufacturing Digital Twin Modeling and Machine Learning Ontology
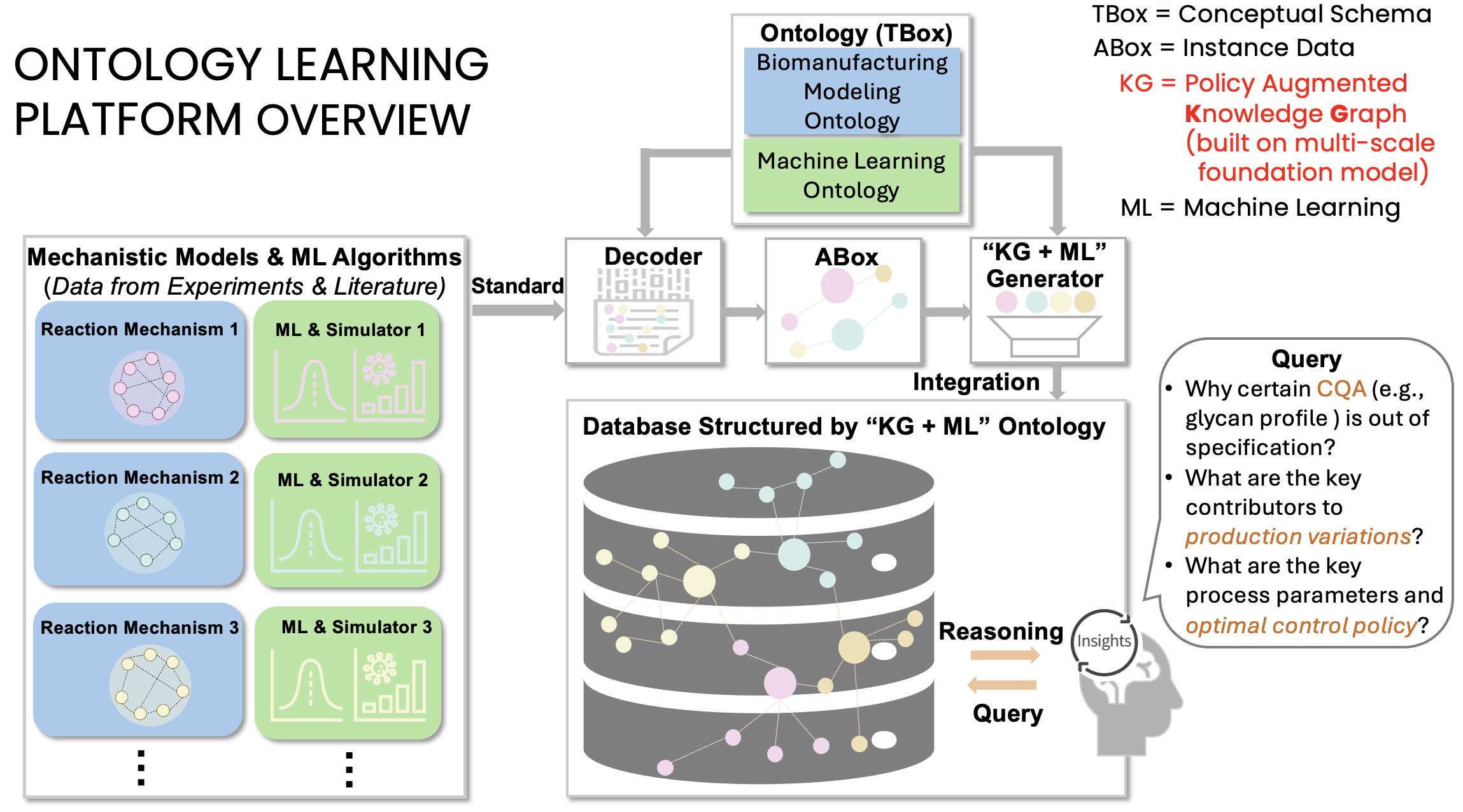
Highlights of a NIST Ontology Project (ongoing):
-
Problem Statement. Biopharmaceutical manufacturing faces critical challenges, including high complexity,
limited data, and fast evolution of biotech (e.g., new bio-drugs and sensor technologies frequently coming to the market).
-
Technology Development. To address the challenges and create a unified digital platform, the objective of this project is to develop
a novel ontology platform that can integrate process data and models, enhance interoperability, and support flexible, optimal, automated and regulated biomanufacturing.
1) We represent bioprocesses as modular Biological System-of-Systems foundation models that are semantically grounded by a domain-specific ontology.
2) By treating molecules as agents and characterizing their causal relations and interaction mechanisms (e.g., regulation, activation, inhibition), the ontology formalizes prior knowledge and serves as a semantic layer for data integration.
3) This structure provides the formal backbone necessary for reasoning and embedding Verification, Validation, and Uncertainty Quantification (VVUQ) workflows directly into the modeling and digital twin development processes.
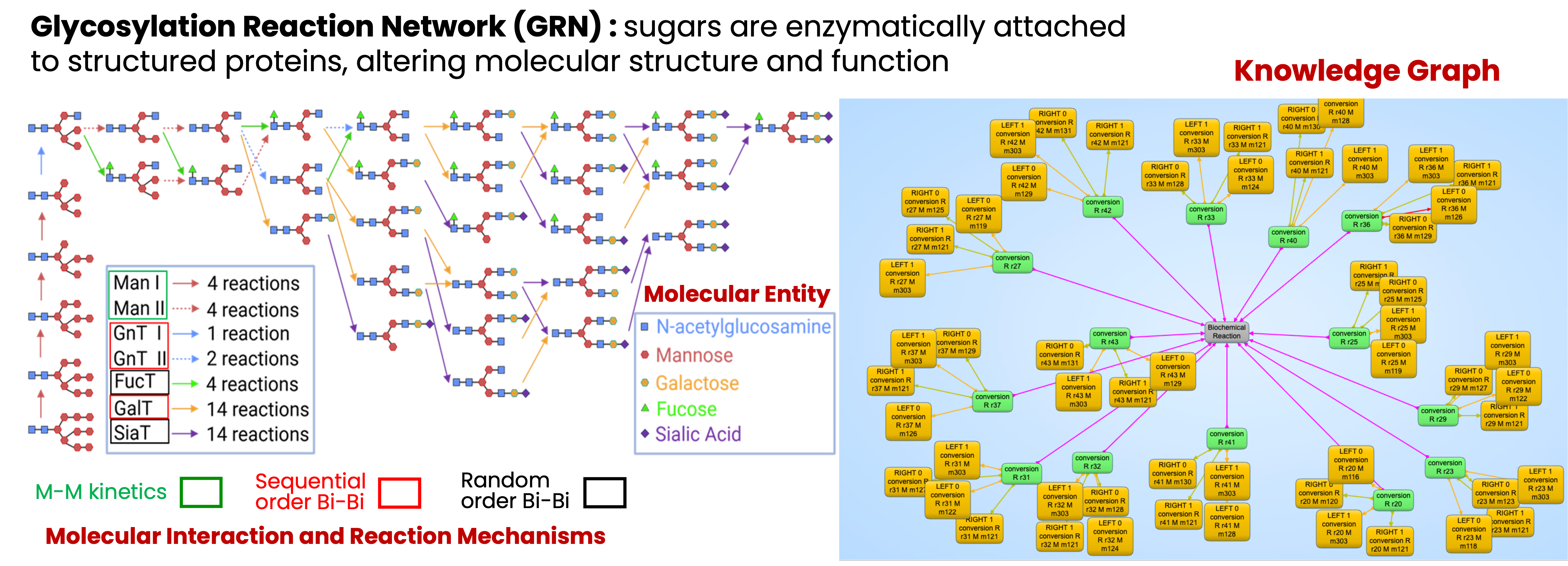
4) We validate this framework through a glycosylation network case study, highlighting how the ontology supports critical quality control, root-cause analysis, and systematic reduction of manufacturing variability, thereby contributing to reliable and credible biopharmaceutical production.
5) Users can interact with our ontology framework through natural language queries, enabling advanced reasoning and deeper scientific understanding.
- Projected Impacts.
This platform paves the way to establishing a unified ontology and general digital twin framework:
1) Support the evolving needs of biomanufacturing domain and enhancing data interoperability and analysis.
2) Enable integration of distributed manufacturing systems, promote data reuse across physical and digital platforms, and accelerate the development of flexible, robust, optimized, and compliant production processes.
Modularized PAT Online Training Platform to Accelerate the Workforce Innovation in Biopharmaceuticals Manufacturing
The video demonstrates interactive interfaces (still under development). It includes
1) A plant-wide simulator generating experimental trajectories of integrated bioreactor cell culture and chromatography purification processes.
2) A Raman spectroscopy simulator taking state concentrations from the plant-wide simulator as inputs to produce Raman spectra.
3) Risk-based ML/PAT training modules including root cause analysis, DoE, integrated bioprocess monitoring, prediction, and robust control.
See the project highlight in NIIMBL 2022-2023 annual report.
Highlights of Project NIIMBL 4.1W
-
Problem Statement. In the project, we create a modularized extensible online training platform on
leading-edge process analytical
technologies (PAT).
It can provide large-scale, low-cost, and high-quality life-long customized training, support workforce innovation, facilitate biomanufacturing 4.0, and strengthen the competitive advantage and leading position of the U.S. in innovative biopharmaceutical development and manufacturing. -
Technology Development. Integrating with professional training certification programs on process control, design
of experiments, and data analytics, we create new PAT training materials and tools including:
1) End-to-end biopharmaceutical manufacturing process hybrid models, which can leverage the existing mechanistic models from each unit operation, provide science- and risk-based production process understanding, and quantify the spatiotemporal causal interdependencies of critical process parameters (CPPs) and critical quality attributes (CQAs).
2) Bioprocess sensitivity and risk analyses, which can guide process specifications and troubleshooting, reduce release times, and support quality-by-design (QbD).
3) Risk-based prediction, which can provide reliable guidance on dynamic decision-making, accelerate process development, and support integrated production process automation.
4) Digital twin-based virtual lab (vLab) and real problem-derived case studies to reinforce mechanistic knowledge, support understanding of bioprocess uncertainties, provide experiential learning, and facilitate problem-solving skills development. - Projected Impacts.
1) The proposed PAT online training platform can enrich the current education programs (i.e., biotechnology, biomanufacturing, machine learning) with risk-based integrated bioprocess modeling/analysis/prediction, interpretable Artificial intelligence (AI), and digital twins, which can facilitate end-to-end biomanufacturing self-learning and automation.
2) The platform can instruct trainees with diverse backgrounds to support their career development in current and future biopharmaceuticals manufacturing.
3) The platform can support the biopharmaceutical manufacturing innovations by: (i) minimizing human errors and defects; (ii) accelerating QbD, process specification, and real-time release; and (iii) facilitating the move to modular, efficient, robust, flexible, reliable, intensified, and automated biomanufacturing.
Optical Redox Probe for Continuous Metabolic Monitoring during Natural Products Bioprocessing
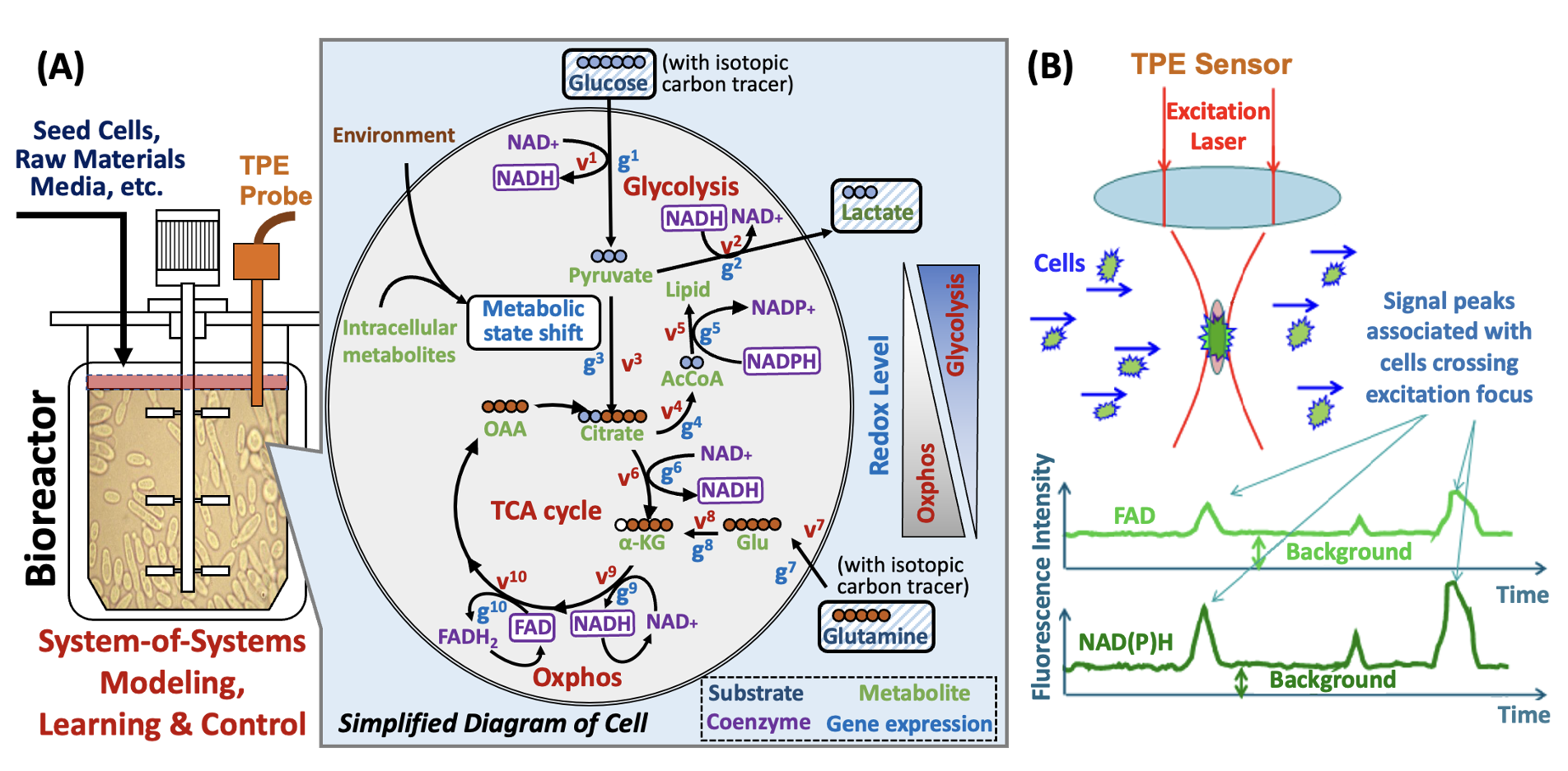
(A) The multi-scale cell culture mechanistic model can integrate heterogenous multi-omics and TPE probe measures to enable the inference of metabolic state of yeast cells, identify intrinsic apoptosis pathways activated in response to hypoxia and nutrient starvation, and guide the selection of bioprocess optimal control strategy to improve productivity and facilitate continuous biomanufacturing. (B) The TPE sensor can provide real-time measurements on intracellular metabolite NAD(P)H/FAD and redox levels that can be used to monitor metabolic state change from glycolysis to oxidative phosphorylation (Oxphos). Since cells are bigger than the focal volume, when a cell flows through the focus, the background emission from other fluorescent components is negligible.
NIH project on Optical Probe and Metabolic Monitoring:
-
Problem Statement.
The objective of this project is to develop and validate a novel optical sensing technology that enables on-line, continuous measurement of cellular-level metabolism of reacting microorganisms and cells in bioreactors.
-
Technology Development.
The proposed two-photon excitation (TPE) fluorescence probe for real-time measurement of important metabolites
within individual cells could guide the development of efficient biomanufacturing of natural products (e.g.,
medicines, biofuels, and foods). We create cell culture mechanistic model and process analytical technology
(PAT) to evaluate the value of the new metabolic data in promoting the development of
effective bioreactor control strategies (e.g., glucose feeding and oxygen control) for improved biosynthesis.
1) Create an interpretable probabilistic knowledge graph (KG) hybrid model characterizing the science- and risk-based understanding of cell culture and fermentation process mechanisms at molecular, cellular, and system levels.
2) This model: a) connects the real-time redox states to the key bioreactor operation parameters (e.g., glucose level, oxygen level); and b) utilizes the cellular-level real-time redox data from TPE probe to improve the prediction of metabolic state change and fermentation productivity.
3) Assess the value of real-time single-cell metabolic data to enhance understanding of biosynthesis and optimize bioreactor performance.
- Projected Impacts. The accurate intracellular metabolism measurement data and the validated PATs provided by this project are valuable for bioreactor operators to select effective feeding strategies, for upstream scientists to develop advanced process technologies, and for microbiologists to discover new metabolism-engineered species and re-engineer the biosynthesis pathways.
Advanced Bioprocess Sensor and Analytical Technologies for Induced Pluripotent Stem Cell (iPSC) Culture Online Monitoring and Automation
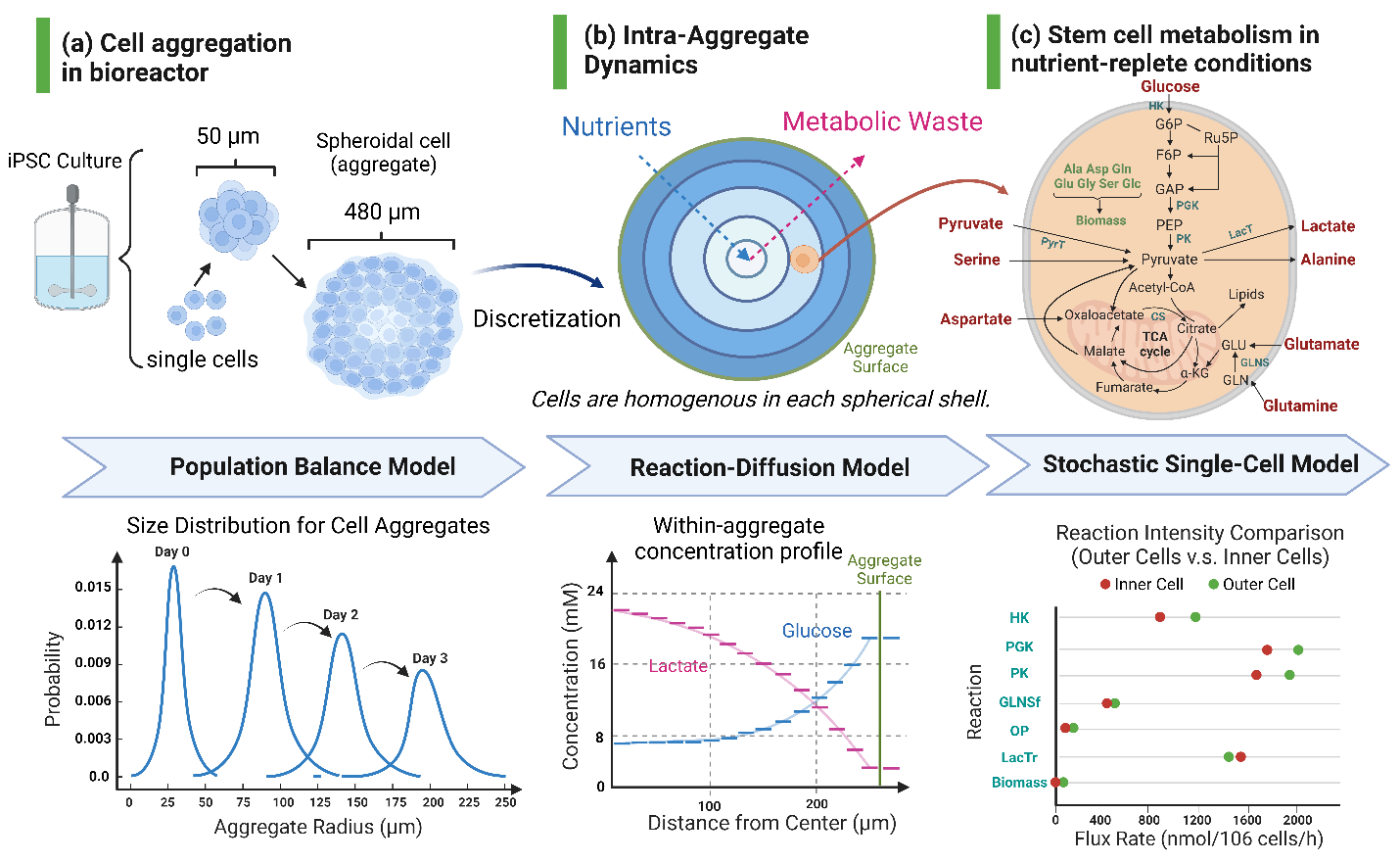
An illustration of our developed Biological System-of-Systems (Bio-SoS) mechanistic model with modular design characterizing iPSC aggregate culture mechanisms. Each cell is a complex system. Many cell aggregates in the bioreactor are Bio-SoS. (a) As the aggregates grow in size, spatial heterogeneity in microenvironment shaped by cell-cell and cell-ECM interactions increases metabolic heterogeneity. (b) A reaction-diffusion model is used to characterize the intra-aggregate diffusion dynamics of nutrient and metabolite concentrations. Cell aggregates are spatially distributed into concentric shells. (c) The single-cell metabolic kinetic model describes cell response to microenvironmental variations. Metabolic heterogeneity is characterized by the difference in fluxes between exterior cells and inner cells locating at different positions in aggregates. In addition, a spatial variance analysis on the Bio-SoS is derived to quantify the impact of aggregate size on cell product quality heterogeneity.
Highlights of Project NIIMBL 5.2T:
-
Problem Statement
1) Large-scale production of induced pluripotent stem cells (iPSCs) is essential for cell therapies and regenerative medicines.
2) However, iPSCs productivity and pluripotency are highly sensitive to culture conditions. Subtle changes in culture conditions can lead to stress and result in cell populations with heterogeneous differentiation potential.
3) Our objectives are to develop a robust online two-photon excitation (TPE) fluorescence optical sensor and a multi-scale mechanistic foundation model that can enhance iPSC culture monitoring and control to improve critical quality attributes (CQAs), including pluripotency, growth rate, and productivity. -
Technology Development.
To achieve this goal and facilitate large-scale production of iPSCs, the key tasks include:
1) Our modular system flexibly assembles digital twins for various production processes, enabling seamless data integration and enhancing predictive accuracy for both monolayer and aggregate cultures.
2) The multi-scale foundation model uses 2D monolayer data to reliably forecast outcomes in 3D aggregate cultures, accelerating the scale-up of iPSC production from lab to industry.
3) Refine the two-photon excitation (TPE) fluorescence optical sensor to measure real-time intracellular redox state of iPSC cultures, i.e., key enzymes NADH/NADPH/FAD for energetic metabolism.
4) Gain comprehensive knowledge of the metabolism of iPSCs cultured in both static and bioreactor systems, including aggregate and monolayer formats.
5) Conduct the iPSC experiments to validate the performance of the TPE sensor and predictive power of the KG-ML. - Projected Impacts.
The value proposition of this project includes:
1) Accelerate the transfer of TPE sensor and interpretable KG-ML technologies to industry and workforce training programs.
2) Integrate iPSC manufacturing systems and facilitate intensified, reliable, automated, and scalable iPSC production in bioreactors.
3) Support Good Manufacturing Practice (GMP) development via Quality-by-Design (QbD) and automation.
Advanced FISH Assay and Multi-Scale Kinetic Model to Improve mRNA Vaccine Potency Assessment and Delivery Process Prediction
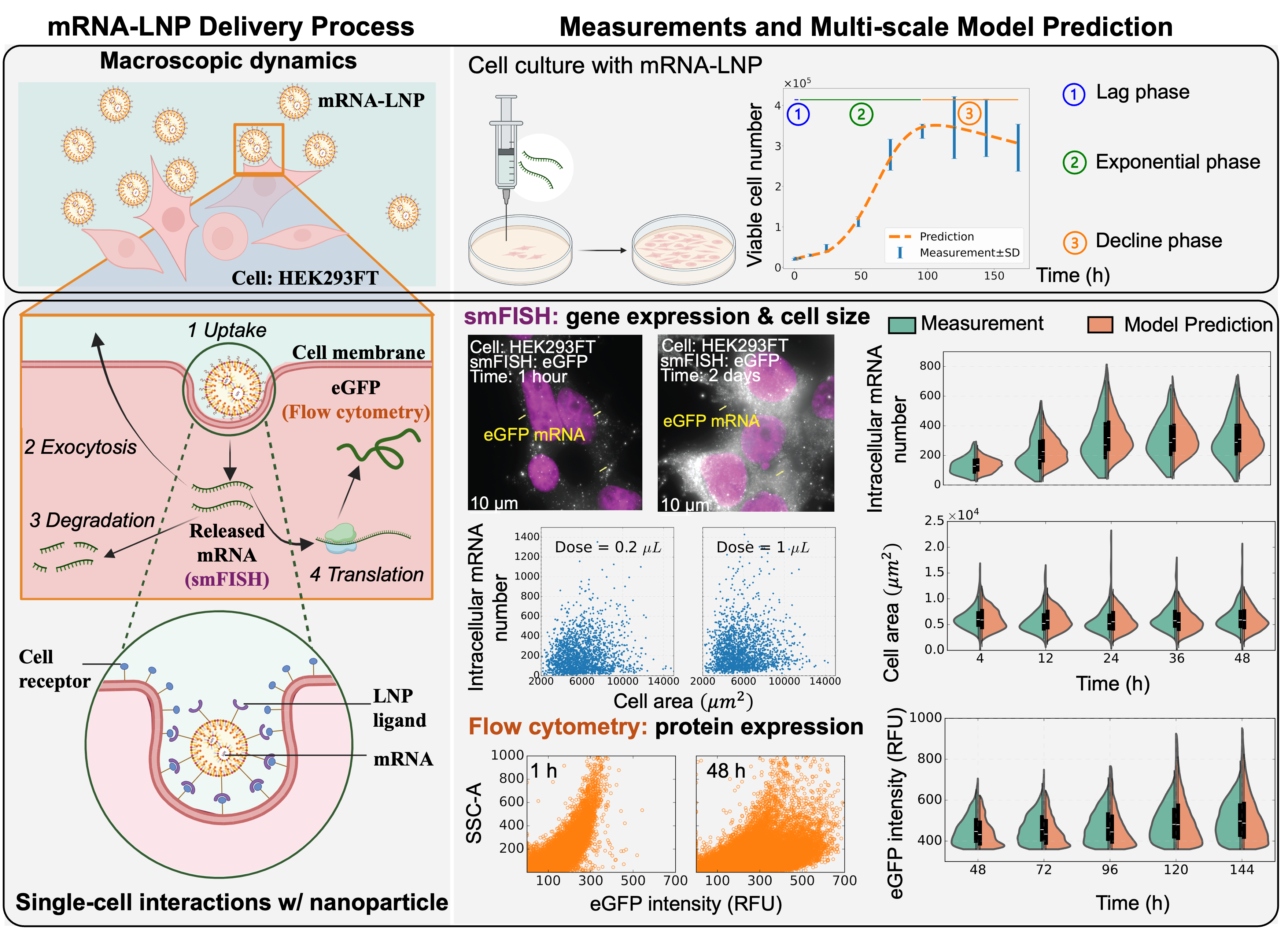
Coupled with advanced gene and protein expression assay technologies, we develop a hybrid model and machine learning approaches to improve the prediction of mRNA vaccine potency and support gene therapy manufacturing quality control. This study introduces an innovative multi-scale kinetics model characterizing the causal interdependence across scales (molecular, cellular, and macroscopic) and accounting for the interactions of nanoparticles and cells. This model characterizes underlying mechanisms on how the dynamics and variations of the mRNA-LNP delivery process depend on critical factors (such as dose, size of LNPs, cell growth and division). Through coupling with advanced multi-omics assays, e.g., single-molecule fluorescent in situ hybridization (smFISH), that can measure the distribution changes of single-cell size, gene and protein expression, the proposed framework can integrate heterogeneous data collected at multi-scale, enable sample-efficient and interpretable learning of fundamental mechanisms of mRNA-LNP delivery and expression, and provide reliable potency prediction.
Highlights of Project NIIMBL 6.1G:
-
Problem Statement
1) The COVID-19 pandemic has promoted rapid development of mRNA vaccines targeting a wide range of infectious diseases. The ongoing emergence of virus variants continuously challenges quality assurance/control (QA/QC) of mRNA vaccines.
2) This enforces the critical need for a multiplexed potency assay and a mechanistic surrogate that can provide a reliable prediction on the potency and quickly screen multivalent mRNA vaccines encoding multiple proteins of variant strains.
3) The objective of this project is to develop and validate single-molecule RNA-fluorescence in situ hybridization (smFISH), mechanistic and hybrid surrogate model for mRNA lipid nanoparticle (mRNA-LNP) delivery and expression processes to ensure vaccine efficacy. -
Technology Development. We focus on improving mRNA-LNP potency prediction and the project includes three key tasks:
1) Create a mechanism-informed, multi-scale kinetic modeling framework that quantitatively captures the coupled dynamics across particle-level, cellular, and macroscopic scales.
2) Develop molecular dynamics and mechanistic models for mRNA stability and translation processes, incorporating RNA folding/binding, structure-function correlation, and molecular interactions (i.e., RNA-to-RNA, RNA-to-protein, RNA-to-lipid, RNA-to-ion).
3) Conduct systematic experiments to validate the predictive power of smFISH and multi-scale kinetic model on mRNA-LNP potency, including tracking mRNA structure-function integrity, detecting mRNA degradation, measuring the efficiency of mRNA delivery and antigen protein translation.
- Projected Impacts.
The proposed smFISH assay, surrogate, and PATs can support QA/QC of a wide range of multivalent mRNA vaccines
and improve public health, including in low- and middle-income countries:
1) Improve mRNA vaccine stability and delivery process consistency.
2) Reduce waste during transportation and distribution.
3) Form a quick response to new virus variants and ensure mRNA efficacy.
4) Accelerate the QA/QC screening for multivalent mRNA vaccine potency.
5) Enhance the predictive power on the potency of new mRNA vaccines and the immune response of people with different genetic backgrounds.
A Modular Mechanistic Model and In Silico Platform for In-Vitro Transcription Process Yield and Product Quality Prediction
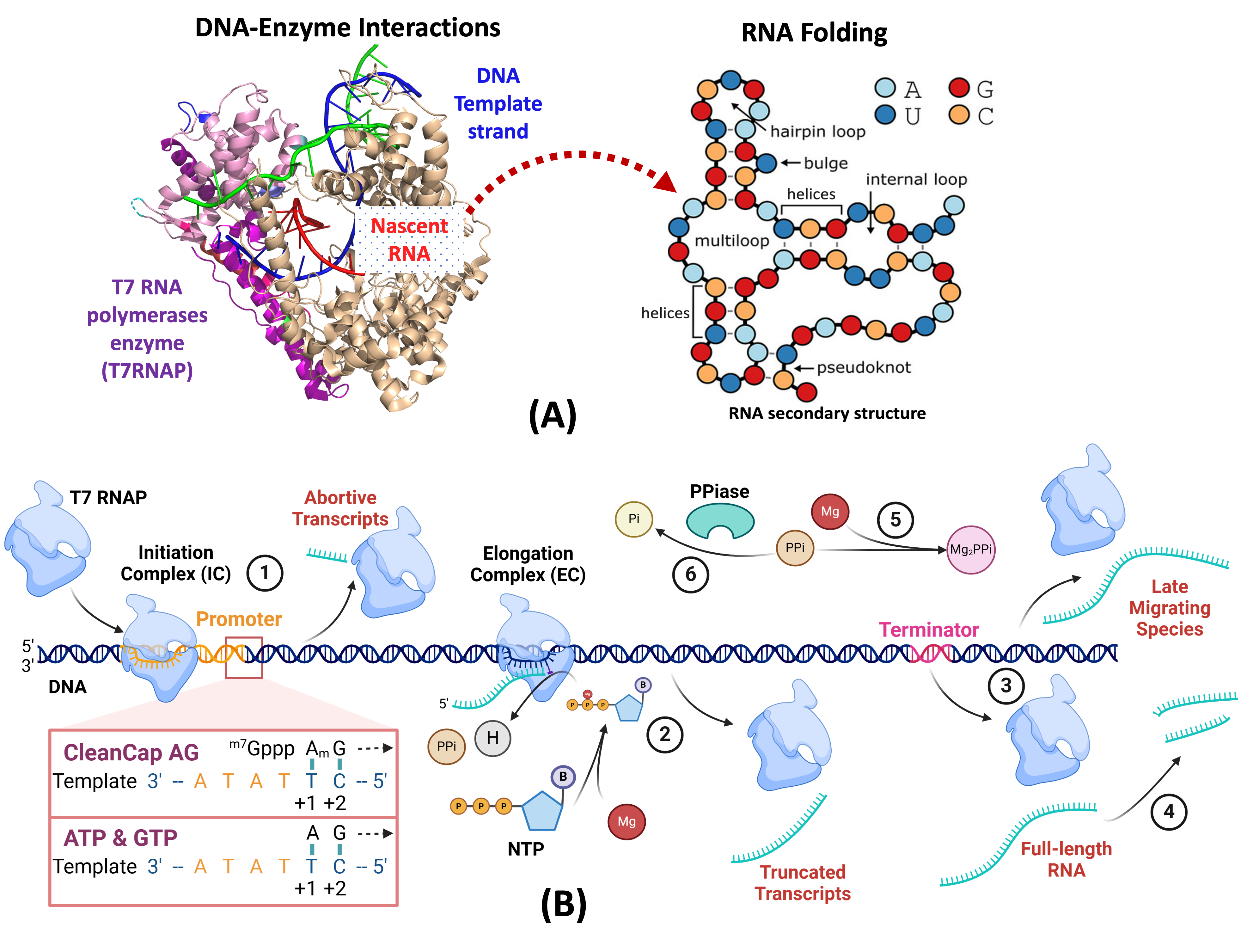
(A) A multi-scale hybrid (mechanistic + ML) model and the digital twin characterize the underlying mechanisms of In Vitro Transcription (IVT) process to improve the estimation of enzymatic reaction rates and the prediction of production processes accounting for the impact of molecule folding/binding and structure-function on molecule-to-molecule interactions. (B) An overview of the molecular components and reaction pathways in the IVT network (Created with BioRender.com) including the key steps: 1) Initiation, Capping and Abortive Cycling; 2) Elongation and Truncation; 3) Termination and Read-through; 4) mRNA transcript degradation; 5) Mg2PPi Precipitation; and 6) Enzymatic degradation of PPi.
Highlights of a Digital Twin Project for mRNA Vaccine Manufacturing
- Problem Statement. To support a rapid-response mRNA vaccine manufacturing, we create an integrated digital platform enabling the fast optimal design of production processes and speeding up the Design-Build-Test-Learn (DBTL) cycle for producing various mRNA products.
-
Technology Development. We create a modular mechanistic
model for In-Vitro Transcription (IVT) process yield and product quality prediction that can accelerate
the development of flexible robust intensified automated mRNA vaccine manufacturing processes.
1) We create a mechanistic model with a modular design for IVT process to advance scientific understanding of underlying mechanisms and improve predictions.
2) The modular design can facilitate data integration and guide an intelligent reuse strategy to enhance rapid-response mRNA vaccine manufacturing against new viral mutations.
3) Leveraging information from biochemical principles and experimental data, a kinetic model is constructed for each module (e.g., initiation and capping, elongation, termination) accounting for mass balances, molecular complexation, and enzyme activity.
4) These modules are then assembled into a mechanistic model to characterize the complex dynamics and interactions governing IVT process performance.
5) Multivariate residual analysis and Shapley value-based sensitivity analysis, guided by domain knowledge, are applied to iteratively improve model fidelity.
6) Due to the high computational cost of each in-silico simulation run, a Gaussian Process (GP)-based Batch Bayesian Optimization approach is employed to accelerate the search for the optimal parameters, while also enabling the use of parallel computing for model fitting and estimation. - Projected Impacts.
1) Facilitate the integration of heterogenous data and information collected from different mRNA manufacturing processes.
2) Advance the scientific understanding of enzymatic mRNA synthesis reaction mechanisms accounting for biomolecular structure-function dynamics.
3) Speed up the development of flexile, reliable, and automated mRNA vaccine manufacturing processes for a rapid response to new viral outbreaks.
Blockchain-enabled IoT for Agriculture and Supply Chains Regulation Compliance, Safety, Interoperability, and Automation
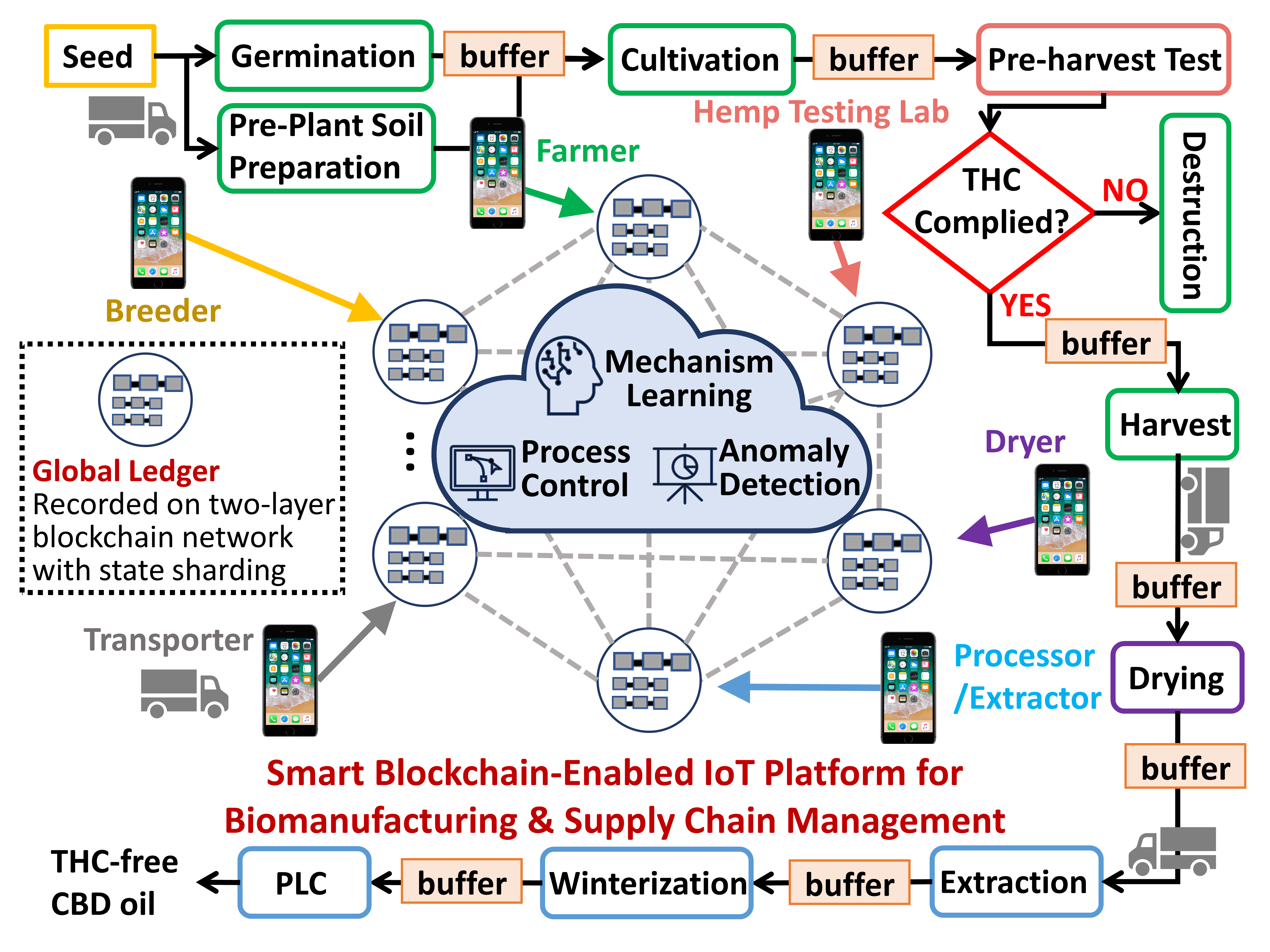
An illustration of a smart blockchain-enabled end-to-end manufacturing and supply chain. Leveraging smart devices, participants can validate and track the information. A two-layer blockchain network deployment can coordinate distributed inspection resources. A regulated and validated global ledger, combined with risk assessment using interpretable AI, can facilitate process mechanism learning, support interoperability with trust, and guide anomaly detection and decision making to enhance yield, speed, efficiency and safety.
Highlights of a Blockchain Project for Regulated Supply Chain Risk Management
- Problem Statement. Our objective is to develop an intelligent blockchain-enabled Internet-of-Things (IoT) platform that can improve interoperability, accelerate agriculture and biomanufacturing industry innovations, and support safe, efficient, responsive, reliable and regulated supply chain management.
-
Technology Development. My research team led on the development of the blockchain platform that was validated during the
real-world small-scale pilot phase 2020 in different states.
1) To address traceability and risk management in regulated supply chains, a blockchain-enabled IoT platform was designed and implemented to enhance transparency, interoperability, and security.
2) The system employs a two-layer blockchain architecture with geography-based state partitioning and hierarchical proof-of-authority consensus, supported by smart contract mechanisms to ensure compliance.
3) A user-friendly mobile application enables stakeholders to collect, validate, and upload real-time data, ensuring end-to-end tracking of materials and products.
4) Simulation and machine learning based decision support tools were integrated to evaluate risk propagation and develop mitigation strategies.
- Projected Impacts.
While demonstrated using an industrial hemp supply chain case study, the developed platform can be applicable to general regulated manufacturing and supply chains in the fields of agriculture, bio-drugs, vaccines, and foods.
Advanced Generative AI for Biological Ecosystems Integrating Multi-Scale Foundation Models, Federated Learning, and Process Optimization
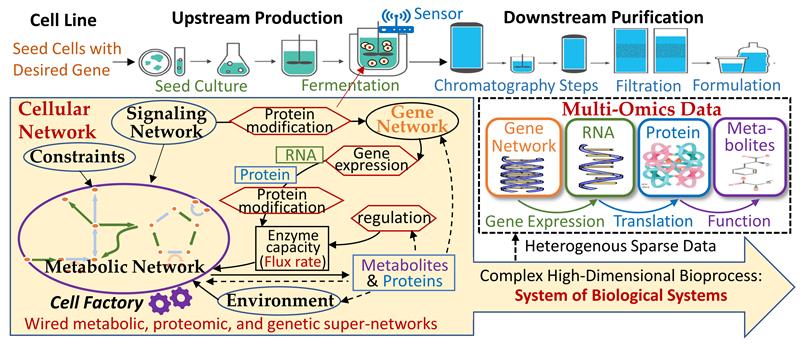
Figure A: With cells (or other living organisms) as factories, biomanufacturing involves Biological Systems-of-Systems (Bio-SoS) with hundreds of biological, physical, and chemical factors dynamically interacting with each other across molecular, cellular, and macroscopic scales and impacting production outcomes.
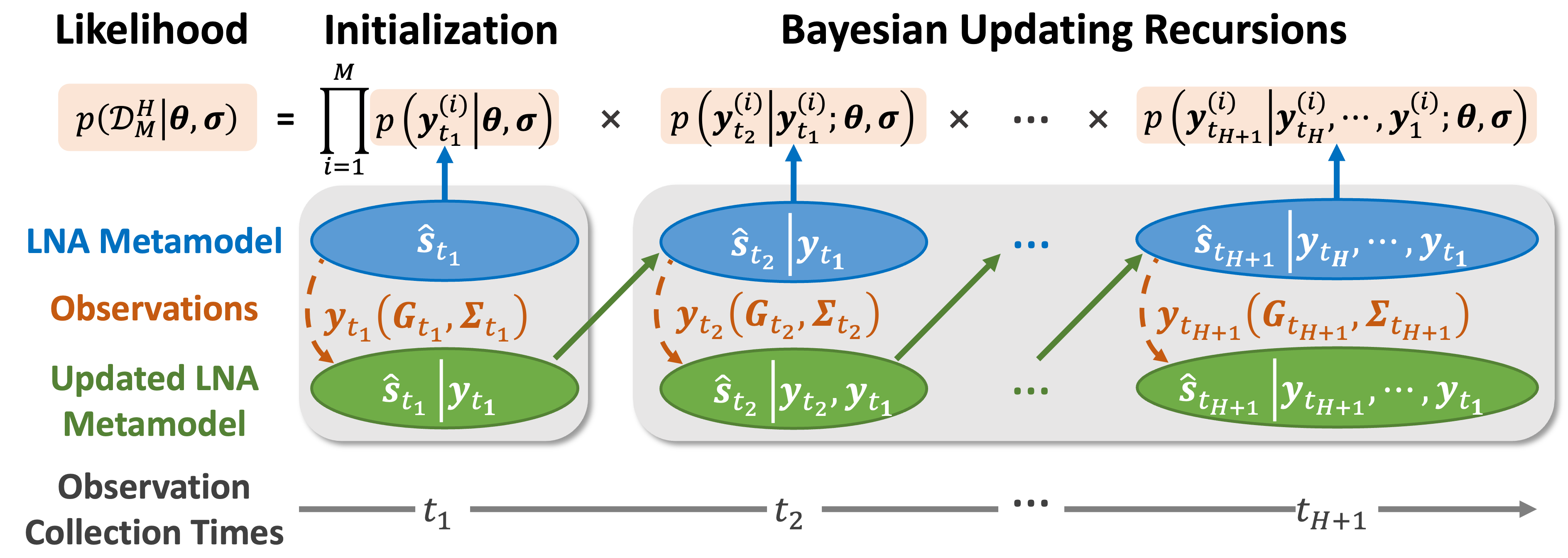
Figure B: To accelerate biomanufacturing systems integration and digital platform development, we create a mechanism-informed Bayesian sequential and federated learning framework built upon a modular, multi-scale probabilitic knowledge graph foundation model formulated as a system of continuous-time stochastic differential equations (SDEs) that quickly fuses sparse and heterogeneous data (y), enabling inference of latent state variables (s) and Bio-SoS underlying mechanisms.
NSF CAREER Award: Mechanism-Informed AI for Bio-SoS to Accelerate Biomanufacturing Systems Integration and Innovations (ongoing)
-
Problem Statement. Our objective is to create a mechanism-informed AI platform on
Biological Systems-of-Systems (Bio-SoS) to facilitate the integration of bioprocesses and
enable a quick assembly of flexible, robust, and optimal biomanufacturing systems.
Since bioprocessing in biomanufacturing is enormously complex and there is a lack of a deep, systemic understanding of underlying
mechanisms, we will answer key research questions:
1) How to create a unified knowledge representation that enables the integration of heterogeneous data collected at molecular, cellular, and macroscopic scales in different production processes from lab to large-scale industrial manufacturing?
2) How to enable sample-efficient and interpretable learning on fundamental mechanisms?
3) How to create efficient design of experiments (DoE) and optimal control strategies within and across different scales? -
Technology Development.
By leveraging advanced sensing technologies, such as optical sensors and multi-omics assays capable of monitoring molecular
and cellular processes, we propose a bioprocess-specific AI framework that fundamentally combines the science of
systems and synthetic biology with uncertainty and intelligence.
1) Create a multi-scale probabilistic knowledge graph (pKG) hybrid (mechanistic + statistical) model with a modular design, as a unified knowledge representation of Bio-SoS mechanisms.
2) Introduce efficient federated learning approaches to fuse sparse heterogeneous data from various production processes and advance the knowledge of input-output causal interdependencies.
3) Construct interpretable digital twin calibration and reinforcement learning on the pKG, accounting for uncertainties due to limited understanding, to guide optimal robust decision making and maximize information gain. - Projected Impacts.
1) Build a world-leading workforce pipeline through a back-propagating strategy that starts with current workforce training and extends to education for college and high school students.
2) Develop a virtual lab and AI training platform with an intuitive interface and scalable, modular backend to enable seamless interoperability from education to research labs to industrial manufacturing.
3) Significantly improve biomanufacturing capabilities and accelerate manufacturing systems integration.